Biography
Interests
Shimon Shatzmiller*, Rami Krieger, Galina Zats, Inbal Lapidot & Ludmila Buzhansky
Department of Biological Chemistry, Ariel University, Ariel, Israel
*Correspondence to: Dr. Shimon Shatzmiller, Department of Biological Chemistry, Ariel University, Ariel, Israel.
Copyright © 2019 Dr. Shimon Shatzmiller, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abbreviations
2_P-AMP – 2_-phospho-AMP portion of NADP+/H;
CPR - cytochrome P450 reductase;
dRf - 5- deazariboflavin;
ET - electron transfer;
Fd - ferredoxin;
Fdox - Fd in the oxidised state;
Fdrd - Fd in the reduced state;
Fld - flavodoxin; Fldox - Fld in the oxidised state;
Fldrd - Fld in the reduced state;
FNR - ferredoxin- NADP+ reductase;
FNRox - FNR in the oxidised state;
kap - apparent observed rate constant;
ket - ET first-order rate constant;
Kd - complex dissociation constant;
NMN - nicotinamidemononucleotide portion of NADP+/H and NAD+/H;
PS I - Photosystem I; WT -wild-type

The use of genetics [1] as a tool to achieve the ultimate goal of realizing biomicro-allergy production is essential for the future of sustainable energy. It will also be an important example of a rewarding symbiosis between genetics and energy when it comes to the useful interaction of living creature and technology, which means a perfect blend of biology and industry. The success of this example can also serve as a major change that changes the game by overcoming the ethical, efficiency and economics questions as well as issues that are important to society. As a final word for this discussion on the basics of bio-allergenic production and changing the mechanism from the perspective of genetic engineering techniques.
That is what should happen

In recent years, our, the humans, relation with the microbial world has reached a turbid phase.
Humans combat microbes, and losing thereby the antimicrobial umbrella, the "miracle" found and used for over one century is becoming cumbersome many antibacterial agents became useless due to the development of microbial resistance. The nosocomial infections are killing many thousands in all healthcare facilities around the world, and the human microbiome is newcomer is the focus of remedial work, hoping that it caused many "incurable" diseases, like Diabetes and neurodegeneration. There is still no remedy, but there is some hope.
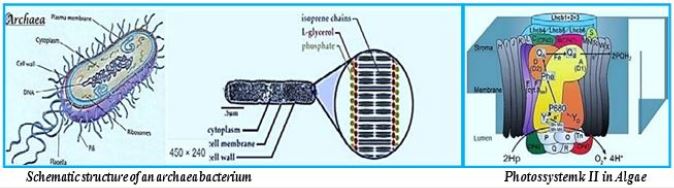
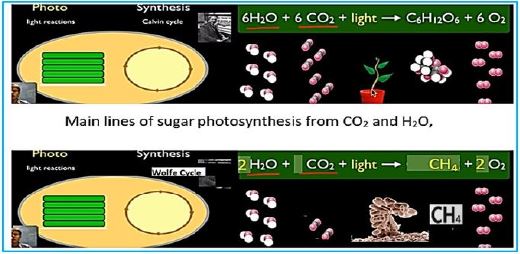
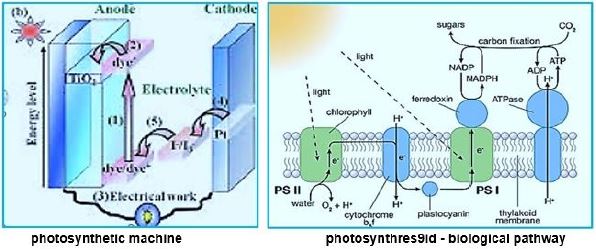
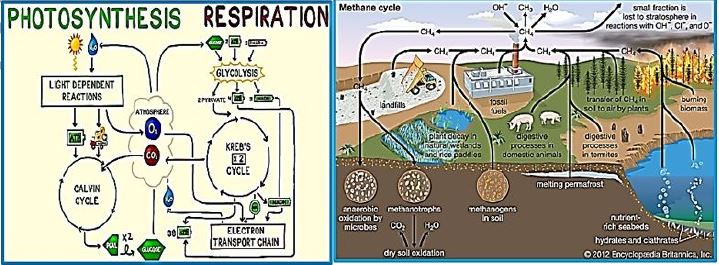
In this dark combat situation, a beam of light shines into the possible harnessing of a microorganism to the effort against global warming. The ability to fixate CO2 by a modified methane-bacterium opens the door to the conversion of the water oxidizing and electron transfer photo-part with the CO2 fixation Wolfe cycle part. The creator managed photo-synthesis by a combination of the enzymatic "photo-oxidation [2,3]" of water and by "light aided electron transfer" into the sugar making machine, the Calvin cycle. It is our task to merge the photo part with the CO2 to methane in the dark part, the Wolfe cycle, to clean our planet's atmosphere from the warming gas CO2 and supply energy by applying the CH4, the product.
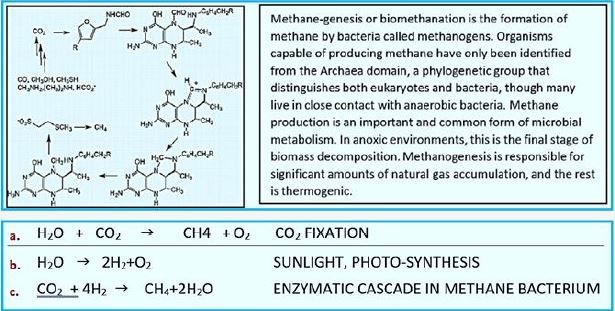
Whereas primitive photosynthetic bacteria such as purple sulfur bacteria and green sulfur bacteria perform anoxogenic photosynthesis, producing elemental hydrogen sulfide with the help of sunlight, cyanobacteria, algae and plants perform oxygen photosynthesis in sunlight and convert water, carbon dioxide into oxygen and sugars. The conversion of solar energy to chemical energy through photosynthesis with oxygen release has evolutionary significance to life as we know it today. In fact, photosynthesis is the only natural process known on Earth to generate oxygen from water. Furthermore, fossil fuels such as coal, oil and natural gas are created.
We may draw parallel lines from the synthesis of sugars from light [4-7], water, and CO2, in the known photosynthesis process to that of the deserved photosynthesis of methane from water and CO2. At the heart of the working for eons, photosynthesis [8-10], the original series of events by which solar electromagnetic energy enters the biological environment, is a fascinating enzyme system that oxidizes water and as a side effect produces the oxygen in the atmosphere. The primary steps of photosynthesis involve the conversion of sunlight into a ‘‘wireless current.’’ In all cases, to form a useful fuel, O2 must be evolved, so it can be released into our oxygen-containing atmosphere and used elsewhere as an oxidation reagent for fuel consumption.
Instead of plant leaves, the bacterial cells are the producer of methane from CO2 and water and light [11,12], where the electron transfer mechanism to NADH and ATP is done in the "photo compartment," and the complicated reduction of the CO2 is carried out by the Wolfe cycle multi-enzymatic reactions and cofactors, in the "stroma equivalent" of the Bacterium.
One of nature's most fascinating and influential enzymes, the water-activating agent photo- oxidoreductase (also known as Photosystem II, a water oxidation compound, and the developing enzyme O2). This enzyme uses solar energy to drive the reduction of plastoquinone initially. From the photosynthetic electron transfer chain using electrons stripped from the water. This reaction is not only one of the significant energy inputs to the biosphere but is also the source of oxygen in the atmosphere...
Main lines of Methane CH4 photosynthesis in engineered bacteria from CO2 and H2O,
We can read in the Scientific American [14] that
Engineered Bacterium Turns Carbon Dioxide into Methane Fuel
If scaled up, batches of bacteria could convert CO2 emissions into fuel, in a single step
Photosynthesis uses solar energy to stored chemical energy by transforming atmospheric carbon dioxide and water into carbohydrates molecules that fuel organisms growth. Scientists have been trying to artificially copy this energy conversion process, with the target of reducing CO2 pollution, And at the same time producing environmentally friendly and sustainable fuels.
Two main approaches are taken to reach the goals,
1. The first is the construction of an artificial catalytic system of many functionalities to reach from water oxidation by light into the fixation of CO2 and production of hydrocarbons or carbohydrates, the gas methane for example.
2. The second is an application of the natural protein-based and cofactors to reach the same goal [15]
Ruminants and Methane Production - Amount Produced
People estimate that the world's Ruminant population [16] produces 77 000 000 tons of methane annually,
which constitutes about 15-18% of all atmospheric emissions [17]. Others speculated that these were
domestic animals responsible for all of the total methane production; [18] Ruminants produce [19] about
97% of the methane produced by domestic animals. About 75% of this production comes from cattle, and
the rest is derived from other animals, such as Water, sheep, and goats. This is given a world population of
some 1.3 billion cattle, calves, and it is assumed that each animal produces 44kg of methane Every year (for
1990).
Methane-bacteria are anaerobic growing by producing methane gas. These bacteria And their exotic metabolism inspired decades of research in microbiology Continues to push the limit of what we know about how bacteria retain energy to it grow. The study of methanogens [20] helped clarify thermodynamics and bioenergetics. The basis of life has contributed to our understanding of evolution and biodiversity, and we have led to an assessment of the social benefit of studying tropical interactions between environmental bacteria since they are essential in the microbial conversion of biogenic methane carbon, high energy fuel. This review discusses the theoretical basis for Conserving energy by manipulators and identifying gaps in the biology of a potential oscillator Being stuffed by organisms that have not been exposed or have not yet been engineered.
Methane and Ruminants
The domestication of animals brought the ruminant animal to human culture, the cattle, sheep, goats,
and camels. These animals support humans with food and utilize that are not suitable for intensive land
cultivation of agriculture, based on plants. However, the ruminants, as beneficial animals, have the drawback of methane production as a by-product in the digestion of grasses and cellulose derivatives. The enormous
amounts of methane that this animal product is vast, as reported below.
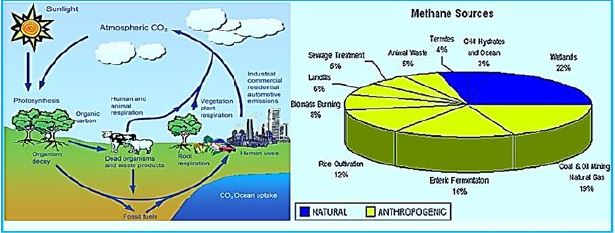
(HINDAWI)Greenhouse gases such as CO2, methane (CH4), nitrous oxide (N2O), and ozone (O3) contribute to climate change and global warming through the infrared radiation in the atmosphere [21]. CH4 is classified as a trace gas and is estimated to have a total global concentration of 1,774 ± 1.8 parts per billion (ppb), with a total increase of 11 pages since 1998 [22]. Methane is a particularly strong gas trace because of its global warming potential, 25 times that of carbon dioxide, and its atmospheric lifetime of 12 years; It is the second-largest anthropogenic greenhouse gas behind carbon dioxide [23]. Methane can also increase ozone in the troposphere of the atmosphere where the greenhouse effect occurs and increase the stratospheric water vapor, which can both add to the radiation power of the gas by about 70 percent. In general, 50-60% of methane emissions are from the agricultural sector, particularly from animal production activities; The primary source of methane is from animal sources from Mazars [24,25].
Domesticated animals, such as cattle, sheep, and goats, produce up to 86 million metric tons (Tg) of methane per year [26]. 18.9Tg are milk visitors, 55.9Tg are beef cattle, and 9.5Tg are goats and goats. Data from Johnson and the department estimate that the annual global contribution of methane is 6.2-8.1, 0.9-1.1Tg of camels, and methane production within the pig and horse axis is 0.9-1.0Tg and 1.7 Tg, respectively.
Methane is produced in ruminants as a product of healthy fermentation of food although methane production can occur in the lower gastrointestinal tract, as in the sex nuggets, 89% of the methane emitted from corpuscles is produced in novels and exhaled through the mouth and nose. [27] When methane inhales into the atmosphere, it has gossiped that the loss of energy derived from food is about 2-12%, depending on a diet [28].
Loss of methane to the atmosphere varies depending on the species. Estimates of energy losses derived from meat, cattle, and beef range from 5.5 to 9.0 percent, 6.0 to 7.5 percent, and 3.5 to 6.5 percent, respectively. For buffalo and camels, the loss of diet energy in the form of methane ranged from 7.5 to 9.0% and 7.0- 9.0%, respectively. Estimates of methane losses from feeders also vary based on geographical location, feed quality, nutrient intake, feed composition, and feed processing.
Methanogenesis
Most of the methane's emissions are directly related to methane-producing methane in warm and humid
soils as well as in the digestive tract of individual animals. Methanogens [30] are microorganisms that
produce methane to produce energy, and they use an anaerobic process called methanogenesis. This process
is used instead of aerobic processes, or with oxygen, because the immune system can not metabolize in
the presence of even small oxygen concentrations. When acetate decomposes in methanogens, the result
is the release of methane into the environment surrounding it. Methanogenesis, the scientific term for
the production of methane, occurs mainly in anaerobic conditions because of the unavailability of other
oxidizing materials. Under these conditions, microscopic organisms called archaea use acetate and hydrogen
to decompose essential resources in a process called fermentation. Acetate Acetate Acetate Acetate Acetate
is produced during anaerobic fermentation to produce methane and carbon dioxide.
H3C-COOH → CH4 + CO2
Hydronotopic Methanogenesis - Archaea oxidizes hydrogen with carbon dioxide to form methane and water.
4H2 + CO2 → CH4 + 2H2O
While atherosclerotic methanogens and hydrogen-hydrogenase are the two main source reactions of atmospheric methane, other original reactions of biological methane also occur. For example, wax found on leaves exposed to UV radiation in the presence of oxygen is an aerobic source of methane [31]. From the beginning, What does it play?
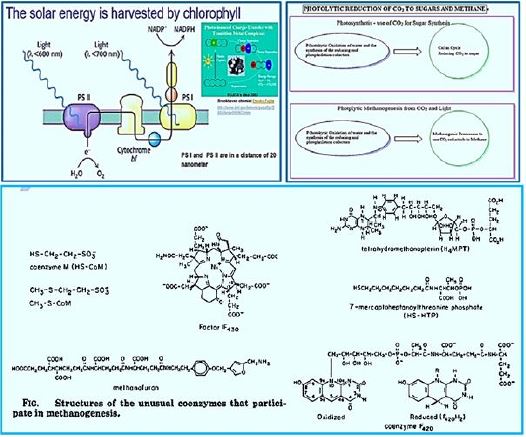
Archaea, which produces methane, can save energy for ATP synthesis (adenosine triphosphate) by producing methane gas. The first indication of methane Biologic gas can be produced by Alessandro Volta in 1776 which discovered flammable Gas and freshwater swamps and the hypothesis that it is stemming from rotting organic matter [32]. It was not. However, by 1933, ideas were first cultivated. Can be found in anaerobic pleasures Especially in low-sulfur environments such as sediments in freshwater pools. Other examples In environments that can be accommodated by animals, they are the digestive tracts of animals (e.g., ants And humans), insects, marine sediments and terrestrial environments. You can isolate from a wide range of thermochemical steps, from acidophilic to alkaliphilic (pH 3.0-10.2), from thermophilic to psycho-profiling (from -2 ° C to 122 ° C), and A master of freshwater to the loophole environments. While melanogenesis was bound to There are no reports of polymicrobial diseases, such as intestinal dysbiosis and gingivitis of methanogens directly involved in pathogenesis through virulence or toxins] [33]. [ To date, methanogens are strict anaerobic archaic and require methane producers. Methanol can grow by reducing carbon compounds in one (C1) [CO2 (carbon dioxide), CO (carbon( Monoxide), methanol, methylamine, and methyl sulfide], acetate or coal to methane through One of several pathways from methanogenesis [34-36].
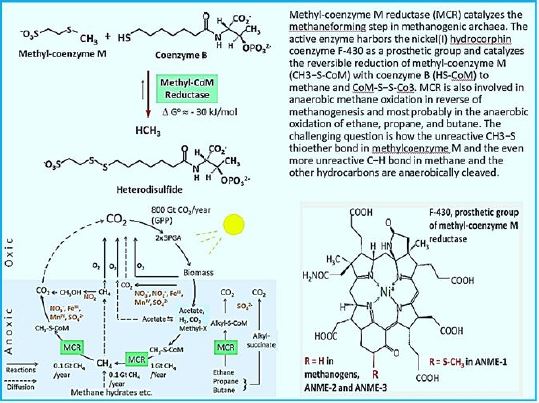
Regardless of the substrate used, methylases M molecule is eventually reduced to methane by methylcoenzyme M reductase Enzyme, Mac [37]. Methanogens differ from bacteria and archaea that may produce Methane as a by-product of metabolism by their binding need to synthesize methane to preserve Energy through the Wolfe Cycle [38,39]. By this distinction, all the known organisms, so far, belong to Euryarchaeal domain. Adaptation and Acclimation of Photosynthetic Microorganisms to in permanently cold environments living microbes [40,41] seems to be a strategy that can permit fixation of CO2 to CH4 using sunlight.
CO2 Fixation to Metha
Omelianski's classic studies [42] on methane fermentationof cellulose werereported in the 1890s. Heisolated
organisms that produced hydrogen, acetic, and butyric acids. He also reported the formation ofmethane
from hydrogen and carbon dioxide:
4H2 + CO2 → CH4 + 2H2O
To achieve sustainable growth in human society, fossil fuels must eventually be altered to renewable resources. Ultimately, the energy and carbon in the synthesized fuels and chemicals must come directly from the sun and CO2. Biological systems promise to speed up the synthesis of such fuels or chemicals. This article discusses the latest advances in the development of CO2 biofuel production processes, which include photosynthetic processes using algae and cyanobacteria and non-photosynthetic electrophoresis [43-48].
Processes using Ralstonia eutropha and other Lithoautotrophi c microorganisms. There exist drawbacks for each of these processes has strengths and weaknesses. While none of these processes have achieved industrial success, the challenges involved may indicate the direction for further improvement within the theoretical possibility boundary. Finally, all biological processes produce a protein-rich cell mass. Ammonium regeneration by diminishing hydrolyzed proteins may close the global nitrogen cycle loop, which is also one of the significant challenges in large-scale biological processes
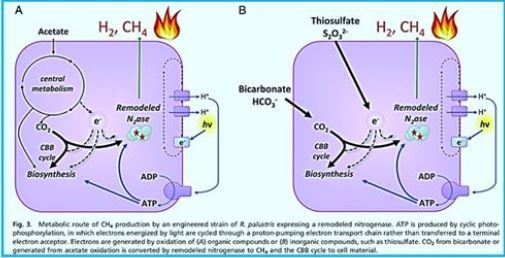
Light-Driven Carbon Dioxide Reduction to Methane by Nitrogenase in a Photosynthetic
Bacterium [49]
Apparently most of us are aware that plants take sunlight and use it to "fix" carbon from carbon dioxide
(CO2) in the atmosphere to sugar compounds in the process of hot-thesis. Natural photosynthesis removes
about 100 billion tons of CO2 from the atmosphere. Every year. CO2's natural release and absorption are
balanced - but Humans release over 30 billion tons a year on top of that, increasing CO2 in the atmosphere
and causing global warming; It exceeds the ability of the plants to remove it. However, what if we could find
a way to optimize carbon fixation? Now, a team of researchers has done just that.
The reaction: Photoinduced water oxidation and reduction of CO2 to methane by [H] that originated in the water
2H2O + CO2 → CH4 +2O2 CO2 FIXATION
A future green economy requires developing effective strategies for converting CO2 to multicarbon compounds that can then be used for other purposes. The authors describe a synthetic cycle for sequential CO2 fixation in vitro, i.e. in the laboratory.
If you've learned the basics of photosynthesis in school, you may remember the simple equation that summarizes the process by which water and carbon dioxide are converted into glucose and oxygen by the sunlight energy:
6H2O + 6CO2 + energy → C6H12O6 + 6O2
This is the most wonderful complexity of the process. There are six naturally occurring carbon fixation pathways; This is the seventh, carotenoid co-enzyme known as Bakelite.
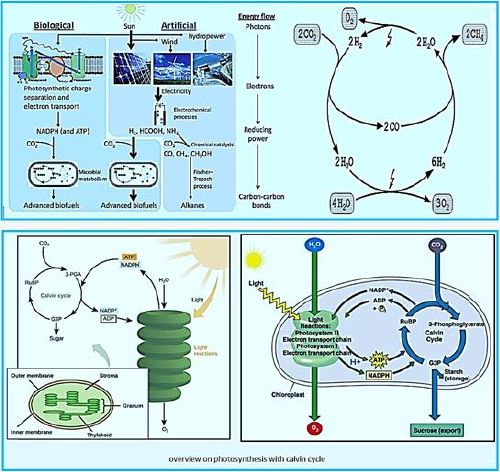
General schemes of solar energy harvesting and carbon sequestration by biological and artificial approaches. Humanmade devices are relatively efficient at harvesting solar energy and reducing electricity generation; While biological biomass meta pathways are more diverse and specific to the synthesis of carbon-based fuels and carbon dioxide chemicals. Recently, a hybrid process has been proposed that combines the artificial "light reaction" with the biological "dark reaction" And demonstrated.
Carbon fixation methanogenic bacteria [50].
Molecule Integrated with Five Carbon Receiving Molecule, Ribulose-1,5-Bisphosphate (RuBP). This phase transforms a six-carbon compound that splits into two molecules of a three-carbon compound, 3-phosphoglyceric acid (3-PGA). This reaction is synthesized by the enzyme RuBP carboxylase/oxygen, or Rubisco.
A simplified diagram (showing carbon atoms but not full molecular structures) illustrating the reaction flowed by Rubisco. Rubisco connects a carbon dioxide molecule to the RuBP molecule, and the intermediate carbon thus formed breaks down into two 3-phospholyser (3-PGA) molecules. A simplified diagram (showing carbon atoms but not full molecular structures) illustrating the reaction flowed by Rubisco. Rubisco connects a carbon dioxide molecule to the RuBP molecule, and the intermediate carbon thus formed breaks down into two 3-phospholyser (3-PGA) molecules.
Diagram showing the molecular structures of RuBP and carbon dioxide, the six-carbon unstable intermediate formed when they combine, and two 3-PGA molecules produced by the intermediate decomposition. Diagram showing the molecular structures of RuBP and carbon dioxide, the six- carbon unstable intermediate formed when they combine, and two 3-PGA molecules produced by the intermediate decomposition. Reduction. In the second step, ATP and NADPH are used to convert the 3-PGA molecules into three-carbon sugar glyceraldehyde-3-phosphate (G3P) molecules. This step gets its name because NADPH contributes electrons to a three-carbon intermediate or narrows it down to form G3P.
Subsequently, the double phosphate molecule receives electrons from NADPH and is reduced to form glyceraldehyde-3-phosphate. This reaction produces + NADP and also releases organic phosphate.
A simplified diagram of the reduction phase of the Calvin cycle, showing carbon atoms but not full molecular structures. A 3-PGA molecule first receives a second phosphate group from ATP (which produces ADP). Subsequently, the double phosphate molecule receives electrons from NADPH and is reduced to form glyceraldehyde-3-phosphate. This reaction produces + NADP and also releases organic phosphate. Reactions from the reduction phase of the Calvin cycle, showing the molecular structures of the molecules involved. Reactions from the reduction phase of the Calvin cycle, showing the molecular structures of the molecules involved. Regeneration. Some G3P molecules produce glucose, while others need to be recycled to replenish RuBP recipients. Renewal requires ATP and involves a complex network of reactions, which my college biology professor liked to call
In order for one G3P to exit (and switch to glucose synthesis), C, O, will begin the subscription, 2, the final subscription molecules must enter the circuit, providing three new fixed carbon atoms. When three C, O, start subscript, 2, end-of-cycle subscript molecules, six G3P molecules are produced. One goes out of circulation and is used to produce glucose, while the other five must be recycled to replenish three molecules of the RuBP recipient. Summary of Calvin Reactors and Products [51].
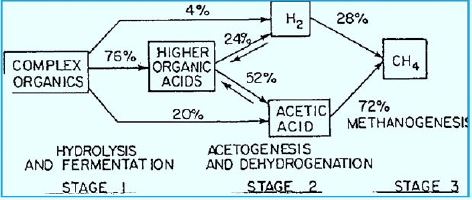
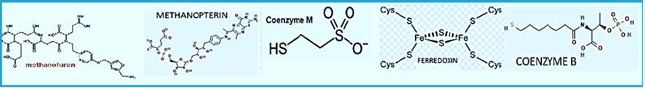
The stepwise conversion of CO2 into CH4 in methane-bacterium (credif ref. [7])
ME, methanofuran; MP, methanopterin; CoM, coenzyme M; Fd, ferredoxin; CoB, coenzyme B.
It takes three rounds of the Calvin cycle to produce one G3P molecule that can exit the cycle and switch to glucose production. We will summarize the quantities of critical molecules that go in and out of the Calvin circuit when creating a single G3P network. In three rounds of the Calvin cycle: 111 G3P molecule goes out of circulation and progresses to glucose production.
Five hundred fifty-five recycled G3P molecules and renewed 333 RuBP accepted molecules.
ATP. 999 ATP is converted to 999 ADP (666 in the fixation phase, 333 in the regeneration phase). The G3P molecule contains three fixed carbon atoms, so two G3Ps are needed to build a six-carbon glucose molecule.
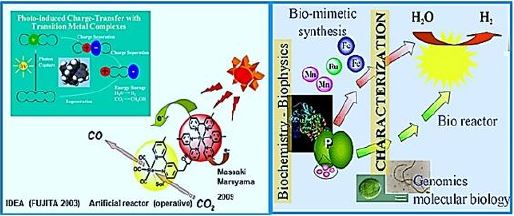
The mn catalyzed Photooxidation of water to hydrogen [52].
Scientists have engineered [1] a bacterium that can extract air from carbon dioxide and make it a single enzymatic fuel. The process draws sunlight into methane and hydrogen production inside the bacterium Rhodopseudomonas palustris, essentially a combustion inverter. These engineered bacteria could guide scientists toward better carbon-neutral biofuels. Researchers published their results [53] in the journal Proceedings of the National Academy of Sciences thebacterium Rhodopseudomonaspalustris, inessence reversing combustion. These engineered bacteria could guide scientiststoward better carbon-neutral biofuels.
The technology of converting CO2 into methane gas (CH4) is at the outset. Methanol-bacterium engineering, which can harness and harvest solar light, and convert the energy in a manner analogous to photosynthesis, appears to enable Wolf cycle to produce methane from CO2, but this has not yet been materialized. We conclude that the process can usually be performed within many of the proposed CO2 storage pool settings. However, extensive fundamental research and development are still needed to define the exact species, environments, nutrition growth accelerators, and the degenerative process economy. As a result, the here presents study does not recommend commercial implementation of Phase III technology at this early stage. This requires, an upto now, unknown living creature: a methane-producing bacterium that can do it by photosynthesis and use CO2 as an essential ingredient [54].
Thye new engineered bacterium should not work in the fuel cell fashion but rather as a photovoltasic cell [55,56]:
A fundamental feature of biotechnology is the use of living things to produce products and solve problems. For example, New Zealand, it has made national headlines through public genetic controversies Different corn, cloned sheep, and animal cell transplantation to the human body. While scientists and government agencies make decisions about their applicability Ethical standards of such research, sometimes the public does not receive full attention decision-making process.
Biological Hydrogen Production
Biological hydrogen production is one of the most challenging areas of biotechnology concerning
environmental and energy problems. Over the past decade, hydrogen energy has advanced on all fronts and
has become en route to all areas of energy. Existing technologies offer high potential for the development of
bio-practical processes for H2 production.
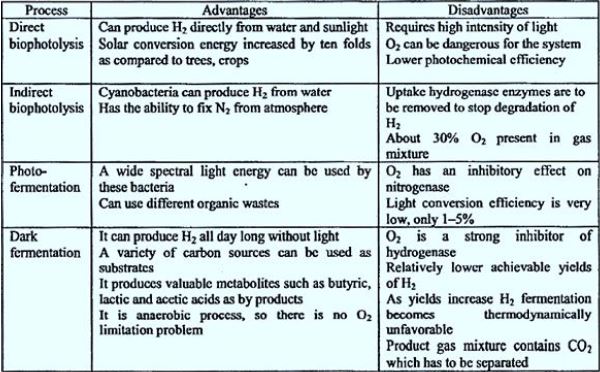
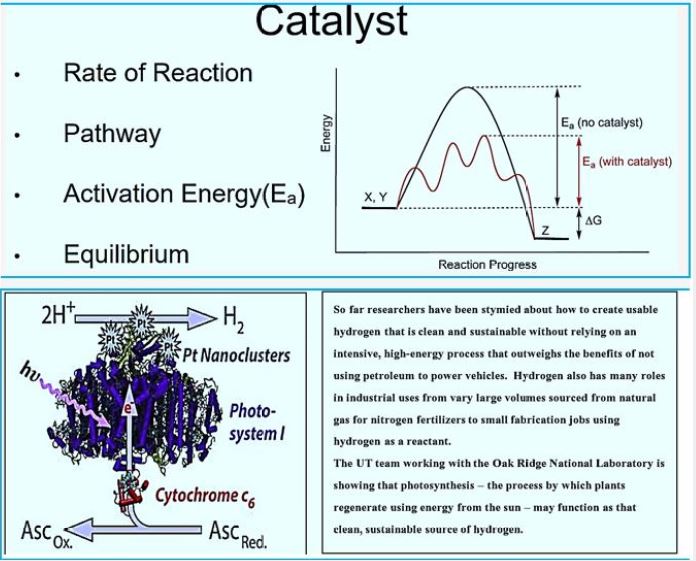
As a sustainable energy source, hydrogen is a promising alternative to fossil fuels. It is clean Eco- friendly fuel. Currently, most of the hydrogen is produced by water electrolysis and by Steam reforming of natural gas. However, biological production of hydrogen has significant advantages On thermochemical and electrochemical. hydrogen. Can be biologically produced by bio-photolysis (Direct and indirect), photographic agitation, dark agitation, a combination of apple and photograph Fermentation and electrolysis in biocatalysis. In this study, there are methods for biological hydrogen production Investigated.
Investigating renewable energy sources is a necessary task in order to replace fossil fuels and reduce the atmosphere pollution. Hydrogen is one of the promising fuels of the future and is seeking further investigation for environmentally friendly ways towards sustainable production. Today, comprehensive processes such as electrolysis and fossil fuels are used to produce hydrogen.
Biological hydrogen production (BHP) exhibits recyclable and economic properties, so it is essential For hydrogen economy. Three basic modes of BHP were examined, including bio-photolysis, agitation in photography And dark agitation. Photosynthetic microorganisms may readily be used as a force for this production sequentially Energy type. Cyanobacteria, blue-green algae (bio-photolysis) and some nonsulfur purple bacteria (photographic fermentation) Utilize solar energy and produce hydrogen during their metabolic processes. Ionic species, including hydrogen) H +) and Electrons) e- (combined with hydrogen gas) H2), with the use of special enzymes, called hydrogens in the case of bio Photolysis, and nitrates accelerate the formation of hydrogen in the event of photo-agitation. However, oxygen The sensitivity of these enzymes poses a disadvantage to bio-photolysis and agitation in photography, whereas the amount of hydrogen The produced substrate unit seems insufficient for dark agitation. This review focuses on innovative advances in the field Research on biological processes, genetic engineering and bioprocessing technologies, such as antimicrobial fuel cell technology, in development Biological hydrogen production.
Today [57], global energy requirements depend mainly on fossil fuels (about 80% of global energy demand). This will eventually lead to the expected depletion of limited fossil energy resignation. Currently, the use of fossil fuel deaths is causing global climatic change mainly due to emissions of pollutants such as COx, NOx, SOx. Hydrogen has the highest energy content per unit weight of any known fuel and can be transported in domestic / industrial consumption by conventional means. H2 gas is safer for dealing with local natural gas. H2 is universally regarded as a remarkably safe renewable energy resource, and an ideal alternative to fossil fuels that do not run counter to the greenhouse phenomenon. The only fuel free from the cab. H2 After oxidation only produces water. can also be used as a fuel for direct burning in an area. In contrast, H2's largest engine is the fertility and oil sectors with, respectively, SO% and 37%. Sales of H2 have risen by 6% annually over the past five years, which clearly refers to increased use of H2 Infinarians as a result of standard fuel quality standards [58].
At present, hydrogen is produced mainly from fossil fuels, biomass and water. The methods of hydrogen production from fossil fuels are
1. Steam refonning of natural gas.
2. Thermal cracking of natural gas.
3. Partial oxidation of heavier than naphtha hydrocarbons.
4. Coal gassification.
Methods of hydrogen production from biomass are
1. Pyrolysis or gassification (which produces a mixture of gases, i.e., H2; CH4; CO2; CO; N2).
2. Methods of hydrogen production from water are
3. Electrolysis.
4. Photolysis.
5. Thermochemical process.
6. Direct thermal decomposition or thermolysis.
7. Biological production.
Generally, hydrogen is produced from natural gas by steam reforming. Other industry methods are coal gasification and water electrolysis. However, these methods use non-renewable users Hydrogen production sources are unsustainable. Therefore, it is necessary to investigate Hydrogen production from renewable energy sources. Biological Hydrogen Processes Production usually operates at ambient temperatures and pressures and is expected to be less Energy-intensive energy from thermochemical methods for hydrogen production. These processes can be used A variety of nutrients as carbon sources. Waste materials can also be used as a carbon source Allowing waste recycling [59]. However, the H2 production rate is low, and Technology for this process needs further development. Clean energy source production and The use of waste materials make the biological hydrogen production novel and promising Access to meet growing energy needs as a replacement for fossil fuels [60]. In this study, Biological production methods are examined.
Biological Hydrogen Production Methods
Biological hydrogen production methods can be classified as below:
1. Direct biophotolysis
2. Indirect biophotolysis
3. Photo fermentation
4. Dark fermentation
5. Two-stage process (integration of dark and photo fermentation)
6. Biocatalyzed electrolysis
This method is similar to the processes found in plants and photosynthesis of algae. In this process Solar
energy is converted directly to hydrogen through photosynthetic reactions (equator (1)). (1)

Algae split water molecules into hydrogen and oxygen by photosynthesis. Generated Hydrogen ions are converted to hydrogen gas by an enzyme hydrogenase. Chlamydomonas reinhardtii is one of the well- known algae that produce hydrogen [4]. There is also hydrogenation activity. The advantage of this method is that the initial feed is water, which is not expensive and Available almost everywhere (Table 1).
A direct bio-photolysis method must force its operation at partial pressure of one atmosphere Of O2, which is a thousand times more than the maximum to be tolerated. Hence, O2 Sensitivity to the enzyme hydrogenase reaction and a pathway for reduction The key problem remains, as it has been for the past thirty years [61].
Hydrogen production by direct photolysis using green algae is currently limited to three Parameters:
(i) The solar conversion efficiency of the photosynthetic mechanism;
(ii) H2 synthesis Processes (i.e., the need to separate H2O oxidation processes from H2 synthesis); And
(iii) Bioreactor design and cost. Several approaches to improve H2 production by green algae are currently
under investigation. These include genetic engineering of light-gathering antennas, Optimizing Light Entry
for Photoreactors, and Improvements to H2 Two-Phase Production systems used in green algae [62]. In
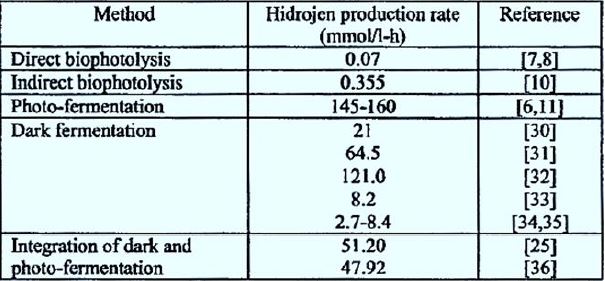
direct bio-photolysis, hydrogen production rates The order of 0.07mm/h per liter has been reported in the
literature (Table 2) [63,64].
Heavy dependence on fossil fuels has caused growing environmental concerns worldwide carbon dioxide release in the atmosphere resulting in global warming. Hydrogen Biological process production demonstrates a promising area for biological energy production Due to its clean, recycled and efficient nature. Existing technologies offer potential Practical implementation, but if bio-hydrogen systems become commercially competitive Must be able to synthesize H2 at sufficient rates to drive fuel cells of sufficient size to do practical work. Further research and development aimed at increasing synthesis and final rates The yields of H2 are essential. If hydrogen's technological potential is realized, it will contribute To the sustainable growth of the global economy by facilitating stable energy supply and by Help reduce future greenhouse gas emissions.
Bacteria Splitting Water
Thanks to the elucidation of the structure and function of a water-splitting catalyst in the Photoshop
II atom-level system, an explanation of the water-splitting mechanism is now at hand. This knowledge
enables the identification of essential criteria for designing similar synthetic catalysts that divide water using
environmentally friendly, low cost and readily available elements. Currently, precious platinum and other
rare metals or metal complexes are used for this purpose. This makes the large-scale production of renewable
energy carriers (fuels) like hydrogen. Very expensive, or even impossible.
Using bio-inspired catalysts, hydrogen or other solar fuel could be produced cheaply by combining solar energy devices with water-split catalysts to produce solar fuels instead of electricity. This will allow the energy economy to overcome the major Solar Power Problems: Sunlight is not available around the clock as an energy source, and electricity is not very suitable for powering motor vehicles. In contrast, the solar fuel concept allows for direct storage of solar energy in chemical compounds, so the use of this energy anytime and anywhere.
For more than three billion years, nature has been using sunlight as its primary source of energy in photosynthesis [65]. During this process, plants, algae and cyanobacteria (blue-green algae) use sunlight to split water and produce energy-rich chemical compounds from carbon dioxide (CO2). The product is carbohydrates (Calvin Cycle), which in nature, act as solar fuels in the living cell. Although the basic reactions involved in photosynthesis have long been known, researchers at the Max Planck Institute for Chemical Energy Conversion in Mulheim an der Roer and the Commissariat à l'Énergie Atomique (CEA) in Skiay, France, have now been able to explain important details of the light-induced water splitting process. As a result, they have refined the scientific basis for producing eco-friendly solar fuels with artificial photosynthesis through sunlight and water, a development that could allow the company to end its dependence on fossil fuels such as oil, coal and natural gas.
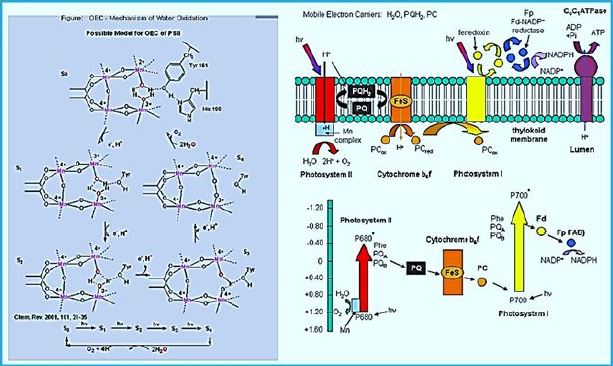
Water Splitting Catalyst
Light-induced catalytic water splitting occurs in a metal complex embedded in an abundant membrane
protein (Photosystem II). This compound consists of four manganese atoms (Mn) and one calcium (Ca)
atom, held together through a network of oxygen bridges (see photo). This compound that oxidizes water
or develops oxygen undergoes a complicated circuit that releases electrons and protons, hence ultimately
hydrogen and molecular oxygen.
In a paper published this week in Science, the German-French research team presents the structure of this manganese-calcium complex directly before oxygen production. This insight into a critical stage in plant photosynthesis is hugely significant: it provides a more detailed understanding of the mechanism involved in photosynthesis and will enable the development of synthetic water-splitting systems based on this model.
The study is the result of close collaboration between the biophysical chemistry and molecular theory departments at the Max Planck Institute for Chemical Energy Conversion led by Wolfgang Lubitz and Frank Neza. Within these departments, Nicholas Cox and Dimitrius Fantasies put together an interdisciplinary team aimed at better understanding the molecular details of water distribution in nature.
Photobiokogical Hydrogen Production
The photobiological hydrogen production process uses microorganisms and sunlight to make water, and
sometimes organic matter, hydrogen. It is a longer-term technological pathway in the early research stages
that has the long-term potential for sustainable hydrogen production with low environmental impact [67].
How Does It work?
In photolytic biological systems, microorganisms - such as green microalgae or Challenges to this pathway
include low hydrogen production rates and the fact that water splitting also produces oxygen, which quickly
inhibits the hydrogen production response and can be a safety issue when mixed in hydrogen at absolute
concentrations. Researchers are working on developing methods that allow microbes to produce hydrogen
for more extended periods and increase the rate of hydrogen production.
Some photosynthetic microbes use sunlight as a driver to decompose organic matter and release hydrogen. It is called protoveratrine hydrogen production. Some of the significant challenges of this trajectory include a meager hydrogen production rate and solar efficiency for hydrogen, making it a non-commercial survey for hydrogen production at this time.
Researchers are looking at ways to improve the collection and use of bacteria to make it more accessible to hydrogen production and to change their usual biological pathways to increase the rate of hydrogen production.

Mixed Catalyst-Bacterial System
Most of life depends on the sun. Through photosynthesis, plants and other organisms harness the sun's
energy to convert water and CO2 into sugars, forming the basis of the food chain. Scientists and engineers
around the world are trying to develop sustainable and sustainable processes Elegant as photosynthesis.
However, it is not so easy to use natural systems as a source of energy. When such organisms are implanted
in bioreactors, the overall efficiency of the photosynthesis obtained is usually quite low, less than five percent.
However, there have been attempts to improve this low efficiency.
Recently, a team of scientists has developed an organic-biological hybrid system capable of driving an artificial photosynthetic process. Their system relies on "artificial leaf" as well as some bacteria to insert carbon fixation into biomass and liquid fuels [68].
Turning the Device On
To evaluate their system, scientists deposited the cobalt phosphate catalyst on a high- carbon surface area
that served as an electrode support. This configuration resulted in much higher currents and much higher
efficiency - the efficiency at which electrons are transferred to the chemical reaction (96 ± 4 percent). The
bacteria were allowed to grow into the cathode interface. When this system was placed in a semi-full batch
reactor in a solution of inorganic salts and organic matter, the reduction of carbon dioxide occurred under
constant stress.
The hybrid system stores more than half of the entry energy in the chemical products of carbon dioxide fixation. They determined that optimum biomass production efficiency (54 ± 4 percent) could be achieved with 36mP phosphate and 2.0 volts over a six-day period.
The biomass output obtained will amount to 180 grams of carbon dioxide captured at 1kW of electricity. If their hybrid device were attached to existing photovoltaic systems, it would have yielded energy efficiency to reduce 10 percent carbon dioxide, surpassing natural photosynthetic systems.
The scientists also estimated the size of this system. They increased the reactor by size and found that the efficiency was not affected.
This process results in biomass and liquid fuel efficiency that is significantly higher than previous integrated bio-electrochemical systems. The energy conversion efficiency achieved in this process is also more competitive with the natural photosynthetic yields.
This process results in biomass and liquid fuel efficiencies that are significantly higher than previous integrated bio-electrochemical systems. Tea energy conversion efficiencies achieved through this process are also more than competitive with natural photosynthetic yields [5].
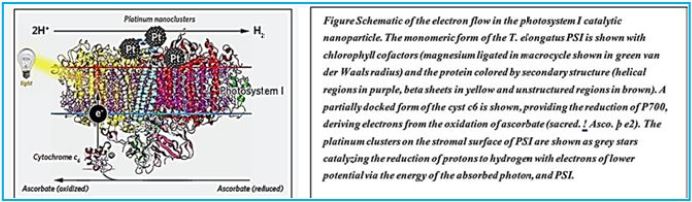
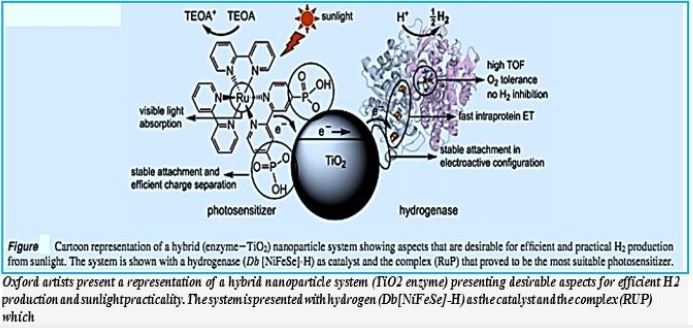
Self-Organized Photosynthetic Nanoparticle for Cell-Free Hydrogen Production
The integrated catalyst [69] design reduces system-level biotoxicity, allowing water- catalyzed catalysis to be
engineered to realize high CO2 reduction efficiencies that exceed natural photosynthetic systems. Because
the Eappl required for water splitting is low (1.8 to 2.0 volts), high ηelec values are directly translated to
high solar-chemical efficiency (ηSCE) when connected to a typical solar-powered device (ηSCE = ηsolar
× ηelec). For a photovoltaic device of ηsolar = 18%, the Co-P | CoPi | R. A hybrid eutropha system can
achieve ηSCE = 9.7% for biomass, 7.6% for bioplastics, and 7.1% for alcoholic fuels. This approach enables
the development of artificial photosynthesis with efficiencies far beyond that of natural photosynthesis, thus
providing a platform for distributed solar production of chemicals.
A De Novo Designed 2[4Fe-4S] Ferredoxin Mimic Mediates Electron Transfer
The interphase to Calvin Cycle
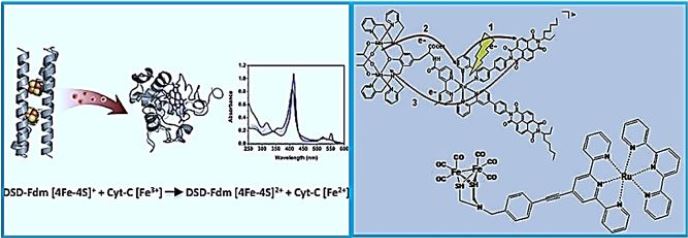
Clusters [Fe-S], nature's modular electron transfer units [71], are usually arranged in chains that support long-term electron transfer. Despite great interest, designing biomimetic artificial systems that simulate multi-negative related proteins, with the ultimate goal of incorporating them into man-made oxidation states, has remained elusive. Here we report an innovative protein from the bis- [4Fe-4S] cluster, DSDFdm, where both clusters are located at 12 Å, compatible with the electronic coupling required for efficient electron transfer. The design utilizes the structural repetition of coil coils as well as the starting scaffold symmetry, the homodimer coil protein (DSD). A total of eight hydrophobic residues in the DSD core were replaced by eight cysteine residues used for cluster ligands [4Fe-4S]. A combination of two clusters [4Fe-4S] focuses on high yield. Both clusters [4Fe-4S] are located in the hydrophobic nucleus of the helical bundle, characterized by different biophysical techniques. The secondary structure of the EPO and holo proteins is conserved; Furthermore, clustering results in the stabilization of the protein concerning chemical data. Most importantly, this de novo-designed protein can mimic the function of natural paradoxes: We show here that reduced DSD-Fdm transfers electrons to cytochrome C, thus producing the reduced stoichiometric.
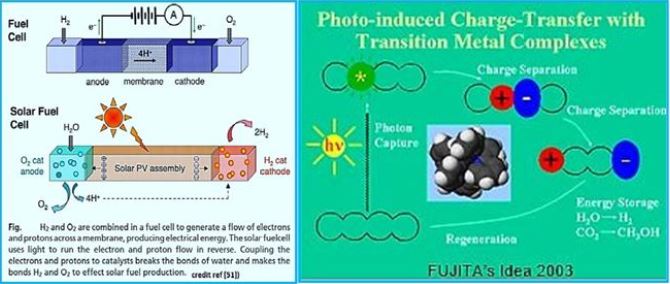
Water + Sunlight = Fuel
Water + sunlight = fuel. [72]. This equation embodies the use of solar energy to tear water molecules into a medium to produce hydrogen, which can be used as an energy-rich fuel for vehicles and power generation. If affordable at a reasonable price, providing a good deal of future global energy demand, which is expected to double between now and 2050. The key to solar water splitting is developing cheap catalysts to efficiently capture and accelerate light the process while reducing the amount of electricity needed to drive electrochemistry. Most catalysts are like that There are far less stellar efficiencies, rely on precious and rare metals or tend to be easily eliminated In difficult working conditions.
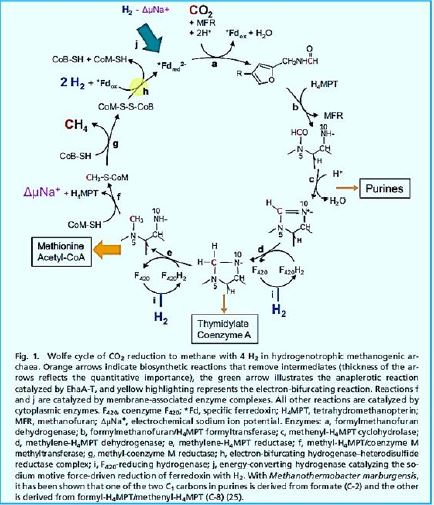
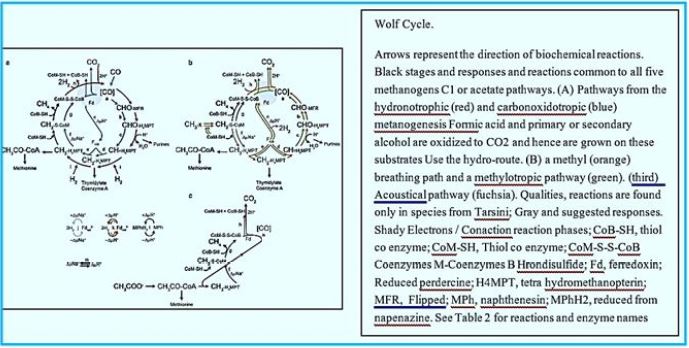
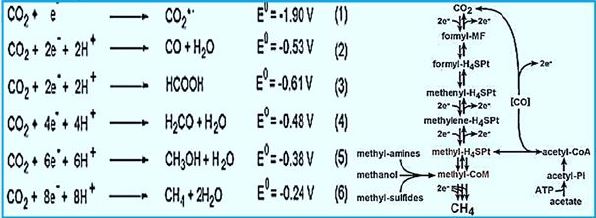
In 1988, Rouvière and Wolfe [74] Suggested methane formation From H2 and CO2 by methanogens archaea can be a cyclical process. Indirect evidence testified that Step one, reducing CO2 to the form of methanol, Somehow he was attached the last step is to reduce the heterodisulphide (CoM-S-S-CoB) Coenzyme M (CoM-SH) and coenzyme B (CoB-SH).
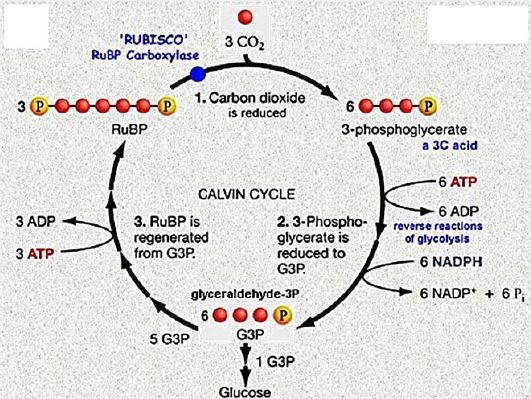
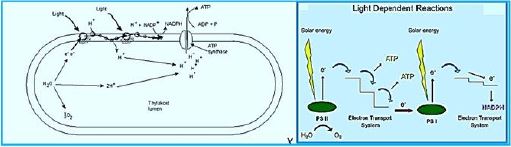
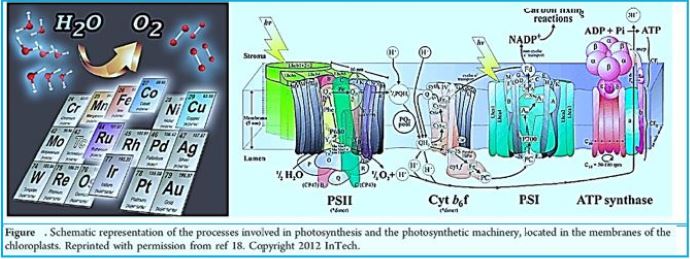
Once the energy from the sun is converted and packed into ATP and NADPH (PS II and PS I), the cell has the fuel needed to build food in the form of carbohydrate molecules. The produced carbohydrate molecules will have a backbone of carbon atoms. Where does carbon come from? The carbon atoms used to build carbohydrate molecules come from carbon dioxide, the gas that animals exhale with each breath. The Calvin cycle is the term used for photosynthesis reactions that use energy that is retained in light-dependent reactions to form glucose and other carbohydrate molecules.
With the help of energy carriers created in the first phase of photosynthesis, the Calvin cycle reactions determine CO2 from the environment to build carbohydrate molecules. An enzyme, RuBisCO, speeds up the fixation reaction, by combining CO2 with RuBP. The resulting six-carbon compound decomposes into two three-carbon 3compounds, and the energy in ATP and NADPH is used to convert these molecules to G3P. One of the three G3P carbon molecules leaves the cycle to become part of the carbohydrate molecule. The remaining G3P molecules remain in circulation to form back to RuBP, which is ready to react with more CO2. Photosynthesis forms a balanced energy cycle with the cellular respiration process. Plants are capable of both photosynthesis and cellular respiration, as they contain both chloroplasts and mitochondria.
In plants, carbon dioxide (CO2) enters chloroplast through the stoma and diffuses into the stroma of the
chloroplast - the site of the reactions of the Calvin cycle where sugar is synthesized. The reactions are named
after the scientist who discovered them and referred to the fact that the reactions function as a cycle. Others
call it the Calvin-Benson cycle to include the name of another scientist involved in its discovery (Figure) [75].
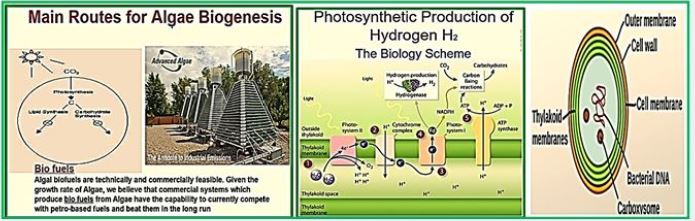
Biohydrogen from Algae
Overview
The Symbiotic [76] and Bionic [77] Biological Systems [78].
Harvesting sunlight is the plant trick that dominated more than eons of years ago, using the sun's energy to feed on the air and water around them in the process we know as photosynthesis. Scientists also figured out how to harness solar energy, using photovoltaic electricity to generate hydrogen that could be used in fuel cells. However, hydrogen could not be an efficient fuel for cars or power generation in a world designed for liquid fuels.
Hydrogen production using micro-algae gets a great deal of observation right away. However, mercantile production of biofuels weighing biomasses of biofuels is not yet usable because of low and high biomass flow processes. Microalgae have been used in the past Four decades in the bio-manufacturing industries of high value-added products and development of useful approaches with environmental applications. The rapid growth rate, the simple growth requirements. The use of sunlight as the primary source of energy is Key Factors for Algae Use. For the last 15 years, considerable progress has been made. The use of microalgae for protein production, nutritional preparations, supplements, molecular tags for the diagnosis and fixation of greenhouse gases [81]. During photosynthesis, the green algae (green algae are probably better for hydrogen production than cyanobacteria whereas the latter uses more energy-intensive enzymes, ATP-requiring nitrogenase for the production of H2 [81,82]). Utilize harvested solar energy to split water, release oxygen to the atmosphere and produce biomass that functions as an excellent food stock At the blue refinery. Green algae are also efficient biocatalysts and can convert solar and carbon energy Dioxide directly for various important compounds, such as vitamins, antioxidants, polymers, And carbohydrates.
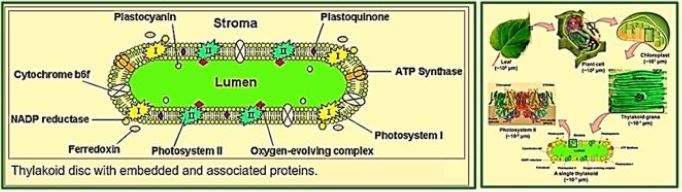
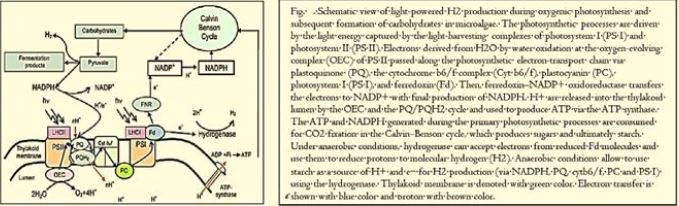
Hydrogen is a non-polluting energy source, which is very renewable and abundant in the universe. Microalgae [84] and cyanobacteria contain the thylakoid [83] membrane. It is a membrane-bound compartment inside the chloroplasts and cyanobacteria. The thylakoid membranes are the site of the light-dependent reactions of photosynthesis. "Thylakoids" consist of a thylakoid membrane surrounding a thylakoid lumen. Chloroplast thylakoids frequently form stacks of disks referred to as grana (singular: granum). Grana are connected by internal or stroma thylakoids, which join granum stacks together as a single functional compartment. Produce hydrogen by decomposing water and organic compounds. For several decades, scientists have known that certain species of algae can produce Hydrogen under anaerobic conditions. Recently, researchers have tried to take advantage of this ability to produce hydrogen that can be used as fuel cells for production electricity - without expensive processes like electrolysis required to split water into it Hydrogen and oxygen [85].
To induce algae's H2 emissions, cells should be placed in the vicinity No oxygen but with access to light. To completely deplete the oxygen supply of the seaweed, The researchers extinguished some of the chloroplast shield needed for oxygen by Adding copper to cells in a closed room. Specifically, the addition of copper Turns off the Cyc6 promoter, which drives the Nac2 gene, which is needed for it Photosystem II (PSII) synthesis. PSII produces oxygen [86].
Scientists have shown that microalgae release hydrogen during the day. Algae generate hydrogen with the help of the enzyme hydrogenase, which breaks down when oxygen is present. Scientists have found three efficient mechanisms to remove oxygen so that hydrogenase can continue to produce hydrogen. According to Jacoby, the discovery of the mechanisms "makes it clear that algae have an unused potential for producing hydrogen fuel."

The staff did not stop there. To further optimize the algae, Thre scientists have modified genetic algae to make the organism produce more hydrogen. They were able to engineer microalgae that produce 400 percent more of the enzyme than regular algae. The goal is to domesticate micro-algae brackish species that can be cultivated to generate hydrogen into hydrogen fuel cells in cars. Scientists have said that one day, even the energy generated by the algae can be used to "drive the wheels of the industry." [87]
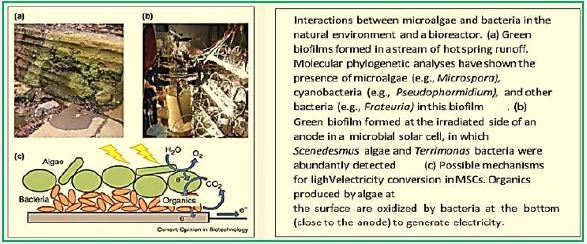
Researchers have developed a technology that could overcome a significant cost barrier to producing cleanburning hydrogen fuel - a fuel that can replace expensive and environmentally harmful fossil fuels.
Cascading innovations, including a brand new one, allowed researchers to reimagine the fuel cell, making it run on methane at lower temperatures. The ruthenium-nickel based catalyst, here in green, was the latest single materials innovation in the new fuel cell. (Image courtesy of Georgia Tech / Liu Lab.) (credit ref [90])
The new technology is a new catalyst that makes platinum almost as well as cost-effective for so-called electrolysis reactions, which use electric currents to split water molecules into hydrogen and oxygen. Rutgers technology is also far more efficient than less-expensive catalysts explored so far.
Twiudrus (Teddy) Aspa, associate professor of chemistry and chemical biology at the School of the Arts, said that hydrogen has long been expected to play a vital role in our future energy landscapes. Moreover, science. That vision for life.
Lichen Symbiosis for Energy Harvesting
Today humans come to the intersection where they can choose a way for the future considering energy
sources and the environment and their practice existence. On the road to a better future, through every
option and every tool will be the key and whatever The alternative should not be neglected. Plunder
Reference, renewables and particular emphasis on Microorganisms like micro-algae are equal Discussions
as a promising energy source.
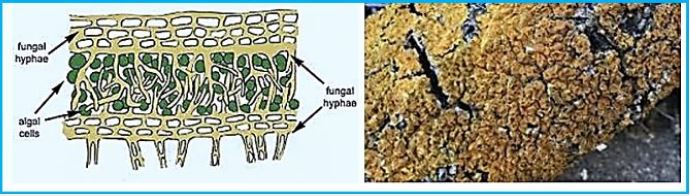
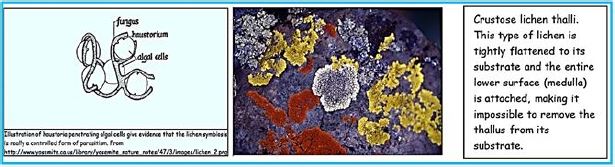
Thallus Lichen which is very thin and flattened on the substrate. The entire bottom surface is attached to the substrate. These lichens are so thin that they often appear as part of the bedding on which they grow. The following link shows a picture of some strong pendants. Brightly colored crustacea usually gives the substrate a "spray- painted" appearance. Thalos has the upper cortical layers, algae, and medals shared by the polio lichen, but he has no lower cerebral cortex. The middle layer is directly connected to the substrate and margins attached to the upper cerebral cortex..(credit ref. [91]).
New fuel research [92] hopes to produce hydrogen efficiently with possible green algae in the future, despite widespread skepticism based on previous research. The study is changing the view of the potential of green algae - and that is good news. The world needs to find a way to generate fuel from renewable energy sources to replace fossil fuels. Hydrogen is now considered one of the most promising fuels for the future and if hydrogen can be produced directly from the sunlight you have a renewable and environmentally friendly source of energy. One of the biological ways of producing hydrogen from solar energy is through photosynthetic microorganisms.
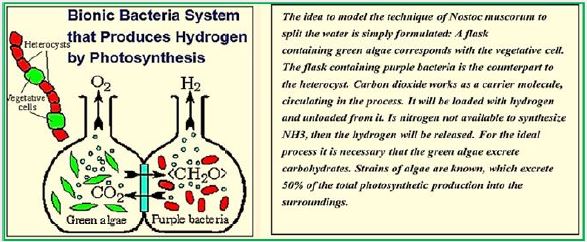
For our purposes, symbiosis will be used herein the most common sense, where there is mutual benefit in the relationship. The two most common examples of fungi are mycorrhizae and lichens, and green algae - purple bacteria for hydrogen production symbiosis.
Symbiosis is usually more scholarly than applicable. In the example of mycorrhizae, you will see that both scientific and applied research have been carried out. Mycorrhiza a symbiotic relationship between plant roots and fungi. The term mycorrhiza literally means root fungus, but in the broad sense of the term, interaction does not always occur only with plant roots, a relationship with mycorrhizal also includes plants that do not have roots, such as psylot and burophytes (moss and heavy liver).
Hydrogen-yielding species of cyanobacteria Cyanobacteria comprise a large and diverse group of oxygen photo-autotrophic prokaryotes, many of which can produce hydrogen . Hydrogen production has been studied in a very wide variety of species and bacterial strains. Hydrogen production occurs in at least 14 types of cyanobacteria, under a huge variety of culture conditions [93]. While a full description of all species and their taxonomy details is beyond the scope of this review, some deserve special mention. Nondiazotropic (Diazotrophs are bacteria and archaea that fix atmospheric nitrogen gas into a more usable form such as ammonia.) cyanobacteria Gloeocapsa alpicola under sulfur starvation shows an increase in hydrogen production [94]. Arthrospira (Spirulina platensis) can produce hydrogen (1μm H2 /12h/mg dry weight of cells) in anaerobic and completely dark state [95]. Another nitrogen-cyanobacterium, Anabaena cylindrica, simultaneously produces hydrogen gas and oxygen in an atmosphere of argon 30 days in mild restricted condition [96] Symbiotic cyanobacteria in the corolla roots of the Cycas revoluta (Cape Sagu) and Zamia furfuracea showed significant uptake of hydrogen in vivo [97], Anabaena sp., Capable of producing a significant amount of hydrogen. Annabana cylindrical nitrogen-starved cells produce an extremely high amount of hydrogen (30ml of H2 / lit per hour culture). Cyanobacteria lacking hydrogen Nostoc punctiforme NHM5 when incubated under high light for a long time, until the CO2 deteriorated culture shows an increase in hydrogen production [98,99].
Cyano-Bacteria is of particular importance because of their great ecological importance in global carbon, oxygen and nitrogen cycles, as well as their evolutionary importance to plants. Photosynthetic cyanobacteria include chlorophyll A and carotenoids in addition to some unusual accessory pigments called filiculins. The blue pigment, picocyanin and red, picuarythrin, absorb wavelengths of light for photosynthesis that the chlorophyll and carotenoids miss. Within the cytoplasm of cyanobacteria there are many layers of membranes, usually parallel to each other. These membranes are photosynthetic thylakoids similar to those found in chloroplasts, which in fact correspond to the size of the entire cyanobacterial cell. The major cyanobacteria storage product is glycogen, and glycogen inclusions can be seen in the cell cytoplasm. Cyanobacteria were thought to give rise to macrophage chloroplasts during the evolutionary events of endosymbiosis. In biochemical detail, cyanobacteria are particularly similar to red algae chloroplasts (Rhodophyta) [100].
The planktonic cyanobacteria determine a huge amount of CO2 during photosynthesis, And "primary producers" are the basis of the sea food chain Environments. Their kind of photosynthesis, which uses the photo system II, Generates a significant amount of oxygen found in the Earth's atmosphere. Since Many cyanobacteria can repair N2 under certain conditions, they constitute the most Prokaryotes with significant nitrogen fixation in free life. Cyanobacteria performed a plant type Photosynthesis (oxygen) at least a billion and a half years ago ,The emergence of plants and the cyanobacteria are supposed to be evolutionary Antecedents of modern plant chloroplasts. Phototropic group Prokaryotes, called prochlorophytes, contain chlorophyll a and b but do not contain phycobilins.
Proclorophytes, therefore, are similar to two cyanobacteria (because they are They are prokaryotic and contain chlorophyll a) and the plant chloroplast (because they are Contain chlorophyll B instead of filiculins). Prochlorone, the first prochlorophyte It has been discovered, phenotypically similar to certain chloroplasts and it is The leading candidate for the type of bacterium that may pass Endosymbiotic events leading to the development of plant chloroplast.
Lithotrophs Lithotrophy, a type of metabolism that requires organic compounds as energy sources. This metabolism is well established in both archaea and bacteria. The methanogens 3use H2 as a source of energy, and many of the extreme thermophiles use H2S or elemental sulfur as a source of energy for growth. Lithotropic bacteria are usually Gram-negative species that use organic substrates that include H2, NH3, NO2, H2S, S, Fe ++, and CO. Ecologically, the most important lithotropic bacteria are the glowing bacteria, Nitrosomonas and Nitrobacter that together bring NH3 to NO2, and NO2 to NO3, and the colorless sulfur bacteria, like Thiobacillus, that oxidize H2S to S and S to SO4. Most lithotropic bacteria are autotrophic, and in some cases may play an important role in the initial production of organic matter in nature. Lithotropic metabolism does not last to acriotics (unless a nuclear compartment has lithotropic endosymbiotic bacteria), and these bacteria are important in the biogeochemical cycles of the elements.
Cyanobacteria are also very important for nitrogen fixation. Cyanobacteria provide fixed nitrogen, in addition to fixed carbon, for their symbiotic partners that make up lichens. This improves the ability of lichens to settle in exposed areas where nitrogen deficiency is constant. In some areas of Asia, rice can be grown in the same pads continuously without the addition of fertilizers because of the presence of nitrogen-fixed cyanobacteria. The cyanobacteria, and especially Annabana, appear in association with the small floating water.
Lichen symbiosis: Nature’s high-yielding machines for rooted hydrogen production Hydrogen is a promising future energy source [101]. Although the ability of green algae to produce hydrogen has long been recognized and published in "Nature" (since 1939 [102]) and has been used in a number of biotechnological applications, but has not yet overcome the greatest obstacle, since the O2 sensitivity of the enzyme hydrogenase. In the present contribution, 75 years after the first reporting of algae hydrogen production, utilizing the natural oxygen balance mechanism, we demonstrate high hydrogen yields by lichens. The ticks have been selected as the ideal organisms in nature for hydrogen production because they are made up of microbiota and photobiont in symbiosis. Scientists accepted that the mycobiome and photobiotic oxygen consumption (an increase of COX and AOX proteins from the mitochondrial respiratory pathway and a corpus-pirate PTOX protein) determines the anoxic conditions required for the activation of the phycobiont's hydrogenase in a closed system. Our results supported the above hypothesis, showing that lichens can activate appropriate bioenergy pathways according to the specific incubation conditions. Under light conditions, they successfully use PSII-dependent pathways and independent PSII (D1 protein decrease and parallel increase of PSA protein) to transfer electrons to hydrogenase, whereas under dark conditions, lichens use the PFOR enzyme and the dark fermentation pathway. To provide electrons for hydrogen. These benefits of acne symbiosis combined with their ability to survive in extreme environments (when in a dry state) constitute them as unique natural plants and produce hydrogen that paves the way for future biotechnological applications.
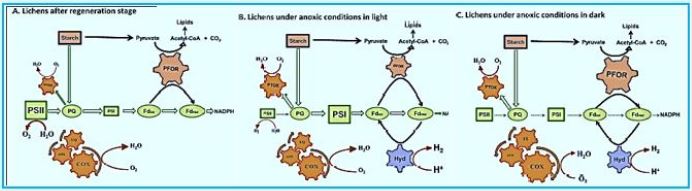
In conclusion, all experiments have clearly shown that lichens can be the solution of nature To overcome the hydrogen-sensitive O2 problem. Polishing can form anoxic Conditions in the system are mainly closed by the consumption of O2 by the mycobiont and at the same time Generate high yields of hydrogen by the photobiont through three different pathways (depending on PSII, Independent fermentation PSII and dark). They have the ability to run Bioenergic pathways are suitable under anaerobic conditions in order to generate hydrogen. Depending on the specific incubation conditions, they can effectively use both induced light Hydrogen production or dark fermentation. The above mentioned benefits of lichens combined Their ability to survive in extreme environments [40] makes them invaluable Future hydrogen production plants that can be used even in space. These traits will bring lichens As very important organisms in the field of bio-hydrogen-energy production. Further investigation Required to pave the way for future sustainable biotechnology Applications, even used on an industrial scale.
It is the partnership - known as a symbiosis - between the various microorganisms that makes them incredibly durable. Algae, and therefore some lichens, can produce hydrogen by splitting water into hydrogen ions and electrons before using a series of enzymes to combine them into hydrogen gas [103].
Photobiological H2 production [104] is an attractive option for renewable solar fuels. It has been shown that Chlamydomonas reinhardtii sulfurless cells produce hydrogen most efficiently among photobiological systems. Scientists investigated the photosynthetic reactions during sulfur deprivation and H fluorescence measurements in production. In the type 6 transition mutant and the transition state 6 (Stm6) of Chlamydomonas reinhardtii.
The incubation period (130 hours) was analyzed at various stages, and changes in the amount and functional state of the Photoshop II (PSII) system were investigated in vivo by electronic and variable paramagnetic resonance spectroscopy.

Hydrogen is a key component in many forms of rocket fuel and the constant production from the durable lichen could help refuel spacecrafts of the future, the researchers say.
Lichens survived Earth's most adverse conditions and the next test of their mettle is to subject them to the harsh environment exclusive to space, such as high radiation levels.
Chemio-Semiotic Gradient
Depending on the substrates, methanogens can obtain between 0.5 and 2 moles of ATP per mole substrate,
which is The lowest theoretical yield except for acetogens grown by acetate production from H2 + CO2 (0.25
moles ATP) and synophases that must be a syntrophic partner to couple metabolism so that the overall
Gibbs The free energy yield for both organisms is positive. There are some key considerations to take If
Gibbs free energy is used to predict methanogen metabolism. The head of these considerations It is that
the predicted energy yields are assumed to be attenuated by chemochemical ATP synthesis. As of today,
everything Methanogens use transmembrane ion levels to produce ATP through ATP synthesis.
Organisms like fermentation Bacteria can use phosphorus at the substrate level to produce ATP, such as in the phosphinulfirovate synthesis. Glycolysis that can be used directly to phosphate ADP to ATP, and releases pyruvate approx. By-product. To date, no such examples of substrate-level ATP synthesis have been demonstrated by methanogen. There are several mechanisms for connecting substrate catabolism to produce a chemio-semiotic gradient [29] Methanogens, such as conjugate transport, scalar translocation, and active suction through membrane-bound oxidoreductocase. Enzymes. Some of these strategies are used for energy conservation antibodies including proton And / or a sodium pumping enzyme complex (Mtr, Rnf) combined with W / Na + antiporter (Mrp), and scalar proton. Transition - place.
Translocation of scalar proton is carried out using the electronic subject methanopenzene (MPh) that uses a nesting loop type and hydrogens to produce a hydrogen cycle (Table 2) [5,68-70]. The use of Gibbs free energy to evaluate methanogen metabolism also assumes 100% ion pumping efficiency For the production of the chemo-mutant gradient. A metabolic reaction may have enough energy to pump four Protons outside the cell, but the chemical bond energy of the substrate may not be perfectly connected to ATP synthesis.
The theory also assumes negligible metabolic entropy at the optimum growth temperature. For example, a The cell may perfectly bind the substrate energy to ATP synthesis, but if the cells have a high rate of ATP consumption For non-growth maintenance, ATP could be consumed quickly without considerable growth. Maintenance energy is defined as the energy flux from a bedding metabolism needed for unit maintenance. Of biomass [71]. Finally, the Gibbs free energy gives a degree of spontaneity / kindness, but in itself cannot Used to predict the rate of substrate consumption or cell growth.
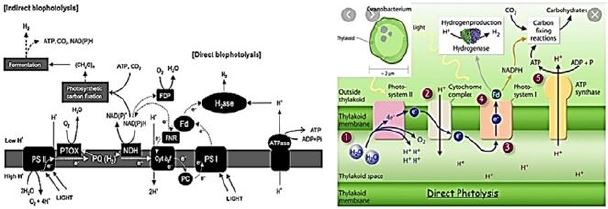
Biophotolysis is a water-splitting process that occurs in biological systems [105,106]. Molecular O2 and
H2 are produced, with light as the energy source. Biophotolysis [occurs in two distinct ways - directly and
indirectly [107-109]. Direct biophotolysis was best investigated in Chlamydomonas reinhardtii microalgae.
This relies on photosynthetic (PSI and PSII) systems and on hydrogen. Light absorption in the form of
photons by PSII (680nm) and / or PSI (700nm) produces a strong antioxidant that can oxidize water to
protons, electrons / electrons, and O2. The electrons reduce protons to form H2. (eq. 1).

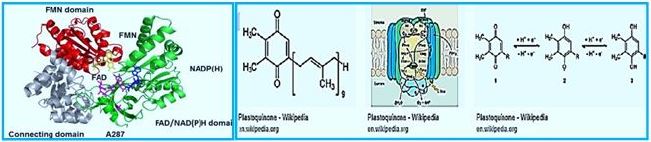
Ferredoxin-NADP + oxidoreductase (FNR) is a flavin adenine dinucleotide (FAD) -containing enzymelinked by a small nuclear gene family in higher plants. It is known that some chloroplast-containing FNR forums are responsible for the final step of a linear electron flow that transfers electrons from Frederoxin to NADP +, while the significant role of FNR in cyclic electron transfer has been discussed for decades. FNR is found from three distinct chloroplast cells (i) in the thylochiode membrane, (ii) in the soluble stroma, and (iii) in the inner chloroplast envelope. Recent in vivo studies have shown that besides membraneassociated FNR, soluble FNR is also photosynthetically active. Two chloroplast proteins, Tic62 and TROL, have recently been identified to form high molecular weight protein complexes with FNR in the thyloid membrane, thus appearing to be FNR's most sought-after molecular anchors for the thylakoid membrane. Tic62 - FNR complexes are not directly involved in photosynthetic reactions, but Tic62 protects FNR from activation in the dark periods.
However, the TROL - FNR complexes affect the photosynthetic performance of plants.
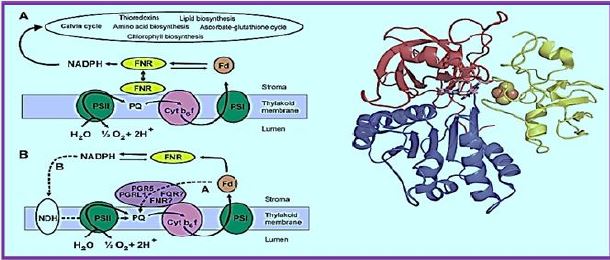
The FAD cofactor is presented as Fd's non-sulfur sticks and cluster as counts
The FNR structure in complex with Fd [15]. The N terminal involved in The FAD binding is shown in red, and the C-terminal domain containing NADP + is shown in blue. The FAD-binding domain consists of a β-barrel structure that is made up of six anti-prolelins Β-strands and covered by α-helix and long loop, whereas the NADP + domain requires Consists of a central parallel β-sheet with five strands surrounded by six α-masses. Ph.D. (Shown in yellow) is associated with the large, shallow rift between the two domains. God Path, electrons are transferred from Fd to the Cyt b6f complex through an uncharacterized part A pathway that includes PGR5 and PGRL1 proteins and possibly FNR as well as hypothetical FQR. Alternatively, electrons may be transferred from Fd to the PQ pool through the NDH dependent A track, which functions in two stages. First, electrons are transferred from NADPH to NDH-1 compound, and second, the PQ pool is reduced by NDH-1 compound. FNR functions in different electron transfer pathways. A. Linear electrons Transfer from the water via PSII, Plastoquinone Pool (PQ), Cyt b6f compound, PSI, Fd and FNR to NADP + are presented as solid arrows. FNR exists as both solubility and bound to thylcoid Shapes.
Reduction of power may also be used to assimilate carbon (Kelvin cycle), amino Acid, lipid and chlorophyll biosynthesis or reduction of active ingredients in redox. B. Possible paths for cyclic electron transfer are presented as dotted arrows. Dependent on FD Chloroplast FNR proteins are molecularly hydrophilic proteins. Weight of about 35kDa. Similarity in the sequence of diverse FNRs Plant species vary from 40% to 97% (see [112]), especially areas Those involved in FAD and NADP + relationships share a high degree of identity. God Three-dimensional structure of chloroplast FNR was described in Many species, with a best resolution of 1.7 Å [8,9,12-18]. The topology of All the FNRs in chloroplast are highly conserved, as is the FNR protein in all studied The species consists of two distinct domains connected by a loop [113], That shows the greatest gender variation. The N terminal The domain (about 150 amino acids) is involved in FAD binding while the The C-terminal domain (approximately 150 amino acids) is mainly responsible for the binding Of NADP + [12] (Figure 2). The FAD binding domain consists of β-barrel A structure built from six anti-paral β-strands and covered by an-helix and Long loop, while NADP + binding area is central A five- stranded parallel β-sheet surrounded by six α-masses. The NADP + - A further binding domain can be further subdivided into two subdomains [114]. God The C- terminal subdomain shows dynamic conformation in daytime stromal A pH that allows increased binding of NADP +, thus effectively functional Control for photosynthesis according to ambient lighting [19].
Molecular hydrogen (H2) exists only at trace concentrations (550 parts per billion) in the lower atmosphere of the earth. However, it plays an essential part in the biochemical cycles of other elements like carbon and is a major component of bacterial metabolism. For example, H2 is an important electron contributor to methane formation in anoxic environments. Here, too, the concentrations are very low (pH 2 <10pa) but the H2 cycle is very high (an estimated 150 million tonnes of biological biomass produced annually in the anoxic ecosystem to fuel methanogenesis) . In anoxic ecosystems, H2's primary role is to transfer electrons between the various participants in the food chain, such as transferring electrons generated by primary fermenters to methanogen Consume H2 Nature has evolved complex metalloenzymes, hydrogenases, which catalyze one of the simplest chemical reactions, reversible oxidation of H2 to two protons and two electrons:
H2 <=> 2H+ + 2e-
Hydrogenases are common in nature and can be found in all walks of life, based on their phylogeny, they can be classified into three separate classes called the metal ions contain.
Biomass production technology can use renewable energy sources like Hydrogen production from biomass, the cleanest form of energy for using by humanity. However, there are major constraints to these commercialization the processes include lower hydrogen yields and hydrogen production rates. To Overcoming these bottlenecks has already been intensively researched on promoting these processes like the development of genetically modified microorganisms; Molecular Engineering of the Key Hydrogenase Enzyme, Development Of two phase processes, etc. The present article examines recent advances Made to this day and also presents the state of the art In molecular strategies to improve hydrogen production.
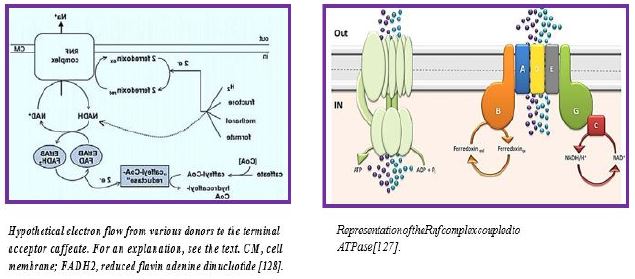
The RNF compound is the only respiratory enzyme in A. woodii. It consists of six unit units that catalyze reduced paroxetine oxidation. The electrons shuffle through iron- sulfur clusters and tablets to NAD + reduced to NADH. Thus, NAD + is the electron acceptor of this anaerobic respiratory chain (see figure below).
This reaction is exergonic (E0 ′ Ferredoxin ≈ -500mV; E0 ′ NAD / NADH = -320mV) and integrates with transcription of Na + across the membrane. The low G0 of the reaction (-34kJ/mol) allows transport of 1 Na + / electron, but the stoichiometry still needs to be determined experimentally. Reversible electron flow. The androgen reduction of paradoxin with NADH as a constrictor is made possible by feeding energy through the Na + River to the cell. In fact, reducing ferredoxin-NμNa + prophylactically is the role of RNF in many aerobic and facultative anaerobic bacteria.
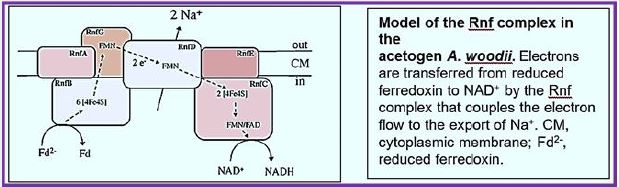
The RNF complex is a prototype of a simple respiratory enzyme, which developed very early in life history, but was preserved during evolution and is presently found in many anaerobic, aerobic and facultative anaerobic bacteria. This is the evolutionary ancestor of the NADH: quinone-oxidoreductase (Nqr) found in some bacteria and is currently being investigated in our laboratory in relation to the electron flow pathway through the enzyme and its mechanistic linkage to Na + transport [115].
The Chemiosemiotic Apparatus [116-118]
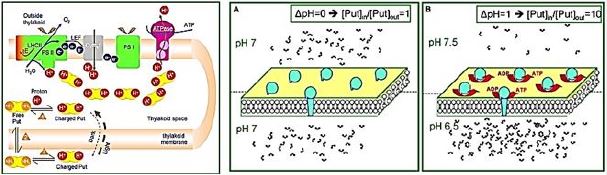
The PA accumulates in the lumen and protects the lumen pH during photo regulation. Photosynthetic reactions produce protons that release vector into the lumen. Lumen oxidation shifts the equilibrium 1 to the right (production of charged lumen). Free deerskin depletion urges new free to disperse from stroma lumen (equilibrium Le Chatelier 2). Finally, the free put in stroma is replaced by the put in the charged stroma (equilibrium 3). The net result of this process (i.e., a new 3 equilibrium point) is ion capture. This accumulation of lumen up to 100 times. The final concentration of Put depends mainly on ΔpH and the countercurrent concentration (like Cl-).
The dual behavior of the Put pool during darkness and light. (A) Equal distribution of protons leads to equal distribution of PA (e.g., at dark). (B) Establishing ΔpH between the two cells leads to unequal distribution of PA. Note the b for which ΔpH = ten times more accumulated in the acid cell.
Aerobic processes, breathing through oxygen as an electron acceptor provides most of the vital energy To maintain cell life. There are facultative bacteria that can use alternative electron receptors, such as NO3-, NO2- And others, but for strict anaerobes, fermentation and / or phosphorus-driven over a preventive gradient are Just means to generate energy. The amount of energy produced is usually between 0.3 and 4 moles Of ATP to a hexagonal mole for absolute anaerobic organisms such as homoetogenic bacteria, Which is far lower than the energy produced by aerobic organisms (up to 38 moles of ATP per Glucose mole). Strict anaerobes have developed energy conservation mechanisms to overcome them This limitation: their ATP output is increased by connecting their metabolic pathways to the generation Of transmembrane ion gradients. This gradient of ions is incorporated into ATPase for ATP production by a Chemio-semiotic apparatus. Some of these mechanisms are well known, such as fumarate A reductase system [123], but others have recently been discovered. Among the newly discovered new energy Conservation systems [3], the energy-converted hydrogen (Ech) link the hydrogen production to ATP production. Another is the Rnf complex, Fredroxine: NAD + membrane oxidoreductase complex This speeds the transfer of electrons from reduced ferredoxin to NAD +. It also provides energy Translate ions across the cytoplasmic membrane, leading to ATP synthesis by a membrane ATP synthase. The Rnf compound (Rhodobacter nitrogen fixation) was first detected in Rhodobacter capsulatus For his involvement in nitrogen fixation [124,125]. The compound is found to be an energy participant Conservation in a number of organisms, such as acetogenic bacteria during autotrophic growth [6]. Breakthroughs relating to the Rnf complex are summarized in the figure below [29,126]:
The anaerobic acetogenic bacterium Acetobacterium woodii straightens a capit (dihydraulic kinemic acid, ethyl (E) -3- (3,4-dihydroxy-phenyl) prop-2-enoate) reduction with hydrogen-derived electrons for ATP synthesis by a chemio-semiotic mechanism [129]. When sodium ions are coupling ions, a process known as coffee breathing. We have addressed the nature of the enzymatic activities that have not yet been involved in this process and their cellular localization. A. woodii cell extract precipitates H2-dependent capit reduction. This reaction is solely ATP dependent but can also be activated by acetyl coenzyme A (CoA), suggesting the formation of capillary CoA before reduction. 2D gel electrophoresis revealed proteins found only in cells grown by Capit. Two proteins were detected using electronic dose-mass spectrometry / ionization spectrometry, and the encoded genes were modified.
These proteins are very similar to the Et (EtfA) and (EtfB) subunits of fluvoproteins to transfer electrons present in various anaerobic bacteria. Western blot analysis of fish that are caused by capit and localize in the cytoplasm. Etf proteins are known electron carriers that transfer electrons from NADH to various acceptors. Indeed, NADH has been used as an electron donor for reducing cytosolic capit. Because the hydrogenase was soluble and used paradoxin as an electron acceptor, the missing link was paradoxin: NAD oxidoreductase. This activity can be determined, and it is interesting to note that it is related to the membrane. A search for genes that could encode this activity revealed DNA fragments encoded to give C and D units of the RNF dehydrogenase Rnf membrane which is a potential Na pump. These data suggest the following electron transport chain: H2 3 paredroxine 3 NAD 3 Etf 3 caffeine-CoA reductase. They also imply that the sodium motive phase in the chain is the reduction of the Nardo-dependent NAD, which is fueled by Rnf.
The electrons flow from PD II to PS I on the "wings of Chemio-semiotic [11]" of many carriers, such as plastoquinone, and receive a further boost of energy, in PS I from 700nm of sunlight. There they come to a super-reducing agent, Frederoxin. They escape from FD to the hydrogen-forming enzyme, where protons reduced to the H radicals that deepen the stable neutral molecule H2 [130].
Green algae and cyanobacteria are photo-autotrophic organisms: they can grow under sunlight and
CO2, without any organic carbon sources. They perform photosynthesis, which converts light energy into
chemical energy according to the Zscheme for linear electron transport (LET). This process, which occurs
in the thylakoid membranes, begins with light absorption by pigment molecules (chlorophylls, picbilins and
carotenoids) that bind to light-picking protein complexes associated with two multi-membrane protein
complexes, Photosystem I (PSI) and Photosystem II (PSII). Pigmented light energy is then transmitted to
pigments in the PSII and PSI reaction center where charge separation occurs. The electron transfer phases
occurring within PSII and PSI. Electron transfer coefficients are arranged in two branches: A and B.1 in
PSII, electron transfer occurs preferentially through branch A, 2 While both branches are active in PSI.
In PSII, the pigment P680 consists of a chlorophyll chloride-related histidine residue. Residues found in
D1 and D2, the two major proteins that form the center of the PSII reaction. Two chlorophyll monomers,
ChlA and ChlB, are located at close distances from P680 on branches A and B, followed by two pheophytin
(Pheo) molecules further away from P680. Until recently, it was believed that PSII cargo separation began in
P680. However, recent experimental observations have led to the suggestion that ++ (or ChlD1) and PheoA
(_0.75 to _0.35 V4,5), whereas P680 is oxidized immediately thereafter (see ref. 2 for a detailed discussion).
To prevent charge combinations between the reducing and oxidizing sides of PSII, fast charge transfer (O300 PS6) occurs between Pheo_ and the plastocoquinone molecule (QA, _0.13 to _.03 V4,5) attached to the D2 reaction center protein. Finally, the electron is transferred to a second plastocovinone (QB, +0.03 V) molecule, which, when reduced twice, leaves its binding site in the D1 PSII protein at the center of the reaction and diffuses through the thylakoid membrane to anchor and reduce intermediate electron acceptors in the cytochrome b6 / f complex. Reduction and oxidation of QB is accompanied by a vector translation of two stromal lumen protons, forming a proton gradient across both sides of the thyloid membrane. The strong oxidant, P680 + (+1.12 to +.27 V; see evidence 4 and 5 for a recent reassessment of this value) An electron extracts from a nearby tyrosine outlet (YZ, +0.97 V) which, in turn, sequentially oxidizes two bound water molecules to the developing O2 complex ( OEC). This reaction releases oxygen and protons to the luminal side of the thylakoid membranes, further adding to the degree of proton.
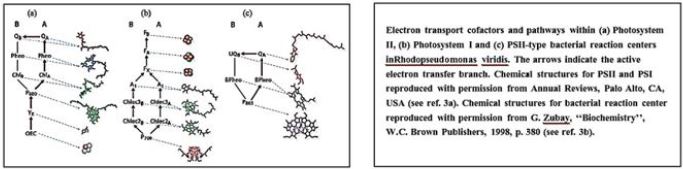
In the context of developing bioprocesses for biofuel and bulk production chemicals, rationally engineered antimicrobial cells to produce such high-yielding molecules and Titers to develop new biological methods that provide economic constraints. Restart Energetic balances of such varieties play crucial roles in performance. Strictly executed processes Anaerobes have a limited amount of energy available compared to aerobic organisms. It Energy is obtained through fermentation and / or phosphorus-driven gradient. such as Anaerobic organisms have developed energy conservation mechanisms to increase ATP yields. It Paper shows the properties of one of these mechanisms paved by the Rnf compound, shows pigeons Membrane complex with ferredoxin NAD + oxidoreductase activity. Rnf compound Ferredoxin electron transfer operation is reduced to NAD + together with ion mover delivery.
Ferredoxin is a common electron carrier for anaerobic bacteria, and with NAD + it is Involved in several pathways for the production of biofuels. This compound was the first Detected in Rhodobacter capsulatus and involved in nitrogen fixation. So it is Be involved in energy conservation in multiple anaerobic creatures, such as acetobacteria Acetobacterium woodii for bacteria that reduce sulfates such as Desulfovibrio alaskensis and autotrophic Bacteria like Clostridium ljungdahlii and Clostridium aceticum. The Rnf compound activates two types Ion transport: It can be sodium or transporter. Both of these shipments form a Gradient of ions, creating a membrane potential then used by ATPase to produce ATP and Thus serving as an energy conservation mechanism. In this review, the information available on the site Rnf compound from genetic organization to its in vivo and in vitro activity in several Summary of microorganisms, with a special focus on the Rnf-protive complex [6,131].
Almost all life forms save energy for growth and development In the process of oxidative phosphorus. It Involves spontaneous, thermodynamically positive transfer Electron from a donor fits through a complex chain of Redox proteins are related to membranes, enzymes and electron carriers, For proper reception. Pumping protons in parallel The surface of the membrane allows energy to be maintained by chemosemiotics Coupling in the form of a driving force (pmf) . The enzyme ATP synthase uses pmf for a proton pair Transfer back across membrane to ATP synthesis from ADP and inorganic phosphate (Pi). Usually higher life forms Use glucose-derived NADH.
Protein-Protein Interactions of Ferredoxin with Its Receptors to Affect Electron
Transfer to Produce Molecular Hydrogen
Protein-protein interactions (PPI) [132] regulate a wide variety of biological processes, involved in almost
every cellular function. Most proteins in living cells interact with spouses and form a complex to regulate
normal functions. PPI employs transport mechanisms, muscle contractions, gene expression regulations,
and signaling pathways [133,134]. PPIs are classified into different types based on their functional and
structural characteristics. According to their stability, interaction, and involvement, PPIs are mandatory and
non- mandatory, gay and hetero, or permanent and transient [135,136].
The general role of light-dependent reactions is to convert solar energy into chemical energy in the form of NADPH and ATP. This chemical energy supports light-dependent reactions and ignites the formation of sugar molecules. The light-dependent reactions are shown in the drawing below. Protein complexes and pigment molecules work together to produce NADPH and ATP.
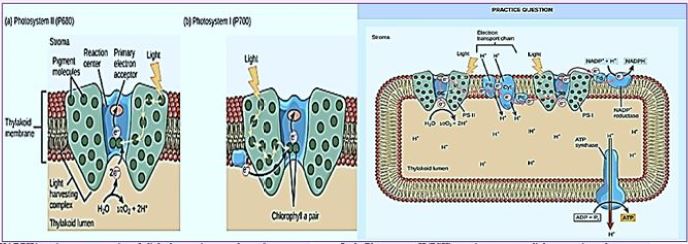
NADPH imaging system consists of a light-harvesting complex and a response center. Pigments in the "Harvest Light" object transfer light energy to two particular chlorophyll objects in the reaction center. The light excites an electron from the chlorophyll coupler, which passes to the initial electron acceptor. The excited electron must then be replaced. In the second photosystem, the electron comes from water splitting, which releases oxygen as a waste product. In (b) the photosystem I, the electron comes from the chloroplast electron transport chain discussed below. (credit ref. [137])
In the Photosystem II (PSII) reaction center, sunlight energy is used to extract electrons from the water. The electrons pass through the chloroplast electronic transport chain to the system I (PSI), reducing NADP to NADPH. The electron transport chain transfers protons across the thylakoid membrane into the lumen. At the same time, water splitting adds lumen protons, and reduction of NADPH removes stromal protons. The net result is low pH in the thylakoid electronic transport chain? A. Water b. Oxygen c. Carbon dioxide d. The Electron transfer [138] reactions (ET) in systems involving proteins require deliberate electron interaction Donates and receives in order to match their redox centers optimally with an effective ET certificate. Such reactions are critical for maintaining the physiological functions of living organisms because they are Involved in necessary actions, such as the photosynthetic ET chain leading to NADPH reduction.

Photosynthetic complex I allow cyclic electron flow around operating system I, an A regulatory mechanism for photosynthetic energy conversion. We report a Cryo- electron microscopy structure at 3.3-angstrom resolution of photosynthetic Complex I from the Cyano-Bacterium Thermosynechococcus elongatus. The model Reveals structural adjustments that allow binding and electron transfer from the Bore synthetic photosynthetic photosynthesis Frederoxine. By mimicking cyclic electron flow with Isolated components in vitro, we demonstrate that Frederoxin directly mediates Electron transfer between the photovoltaic system and complex I, instead of using Mediators like NADPH (the reduced form of nicotine adenine dinucleotide) phosphate). Fixed significant rates for connecting Frederoxin to Complex I Indicates sufficient identification, where the NdhS protein subunit is the Key Component in this process.
The cytochrome b6f [141] complex provides the electronic connection between the photosynthesis system I and the reaction centers of the photosynthesis system II of oxygen photosynthesis and produces a transmembrane electrochemical proton gradient for adenosine triphosphate synthesis. Structure of crystals 3.0 Angstrom of the b6f complex Dion from the Cyano-Bacterium thermophilic Mastigocladus luminous reveals an ample quinone exchange space, stabilized by lipid, which is associated with plastoquinone, a nesting-analog inhibitor, and a novel heme. The core of the b6f complex resembles the analogous cytochrome bc1 complex, but the domain arrangement outside the core and complement of different prosthetic groups are remarkably different. The movement of the outer domain of iron-sulfur protein, essential for electron transfer, must also be different in the b6f compound.
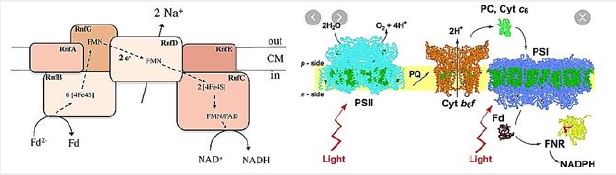
The Rnf complex; [5] is a prototype of a pure respiratory enzyme, which developed very early in life history, but was preserved during evolution and is present in many anaerobic, aerobic and facultative anaerobic bacteria. This is the evolutionary ancestor of the NADH: quinone-- oxidoreductase (Nqr) found in some bacteria and is currently at work, concerning the electron flow pathway through the enzyme and its mechanistic conjugation to Na + transport.
Bibliography

Hi!
We're here to answer your questions!
Send us a message via Whatsapp, and we'll reply the moment we're available!